The agitated detective barks, “I need that DNA now. I have to know if we’ve got the killer in custody. I don’t give a damn how busy you are . . . ” We’ve all been there, on the edge of the couch, watching a cop show on TV and hoping that the magic of DNA testing will give a clear answer to the question “DidHeDoIt?” And indeed, DNA fingerprinting, more accurately ‘DNA profiling’, has transformed crimefighting. If it can be brought to bear, the guilty can confidently be incarcerated, the innocent exonerated, the potential inheritor of the rich father’s will identified, and we can get to resolution after just 42 minutes of drama in CSI. Where did that technology come from, and how does it work?
This post will describe the origins of DNA profiling, how it resulted from the curiosity of a young English scientist, and the confluence of some unexpected discoveries in molecular biology with a driven effort to determine, and to chemically synthesize, DNA sequences. The rapid and unanticipated outcome was that a new technique appeared for the exquisite identification of individual humans by unique patterns in their DNA. The following post goes into the further refinement of DNA profiling by another invention that made it enormously more sensitive. But this, in turn, has led to some issues that raise cautionary flags.
A Mystery in the Hemoglobin Gene
In the mid-1970s, the newly minted English PhD scientist Alec Jeffreys wanted to join an ongoing study of a human gene, the one coding for globin. DNA cloning had just been invented, which had transformed biomedical science. It meant that the DNA of any gene could now be isolated, in pure form, in essentially unlimited amounts. A globin gene was being studied in Zürich, Switzerland. In Jeffreys’ words (1), “No one had ever seen a mammalian gene. No one had any idea of what it would look like.”
Jeffreys joined the Swiss group, and as the work progressed, it became clear that the globin gene had some surprises in store. For one thing, its coding region existed in chunks, with interruptions that apparently contained nonsense. And there were DNA segments lying outside the gene that showed peculiar patterns, consisting of short, tandemly repeated sequence motifs. In the language of DNA, that might look like:
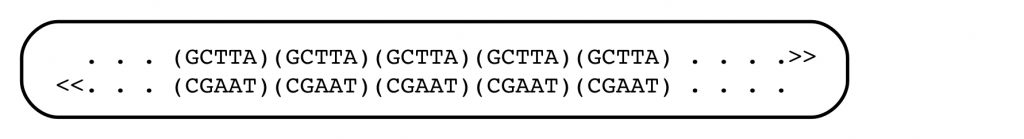
In this case, the short sequence motif GCTTA is repeated five times, head to tail, in one strand of the genome, and its complement in the opposite one. The repeats are inserted into conventional, non-repeated, regions of DNA.
These patterns of tandemly repeated short strings were found scattered throughout the genome, and when Jeffreys took a faculty position at Leceister University in England, he turned his attention to them. He soon found that the patterns of repeated short sequences were highly polymorphic in the human population (from the Greek, ‘many forms’). One person might have 10 repeats of the GCTTA sequence, and another person might have only 7 at the same point in the genome. This polymorphism was in contrast to the highly conserved sequences found in protein-coding regions. The short, tandem repeats were thought to lack a function, since they lay outside the ‘structural gene’, the part coding for protein, and they didn’t make genetic sense. There was no evidence that they were expressed. This polymorphism, of Variable Number Tandem Repeats (VNTR), was the foundation for what became known as DNA profiling.
A Family Re-united
The application of DNA profiling to human affairs began, not with a criminal case, but a humanitarian one. In 1983, Mrs. Christiana Sarbah, of Hammersmith, London, faced a heart-wrenching problem concerning one of her children (2). She had immigrated to England from Ghana a few years earlier, together with her two sons and two daughters. She was estranged from her husband, who stayed in Ghana.
A couple of years after arriving in England with her four children, Mrs. Sarbah’s son Andrew, then 8, decided to rejoin his father in Africa. But five years after that, he wanted to return to his mother in England, and that’s when the difficulties started. British immigration officials weren’t sure he was really Andrew, son of Christiana. Maybe his passport was forged? Maybe he was a nephew? Maybe he was a completely unrelated boy? Any documents his mother could provide, and her protestations, didn’t convince them. At first, the officials wouldn’t even let him leave Heathrow airport, but her Member of Parliament intervened, and Andrew was allowed to live with his mother until an appeal was heard. At this point, the Hammersmith Law Centre, which provided legal aid to the underprivileged, took up the case. But their evidence, including photographs and depositions, did not persuade the authorities either.
There were existing methods for determining relationships. The blood group method, which is one of the best of the older methods, illustrates their shortcomings. There are only four types of blood group substances in the human population. This makes it fairly easy to match blood donors and recipients, but it also gives blood group determination only limited value in identification. As older watchers of Law and Order know, it doesn’t provide a convincing link between a blood sample and a particular person (“… Tell me doctor, how many people with that blood group are there in New York City? …. 2.5 million, you say. And how many of their blood samples, other than my client’s, have the detectives asked you to look at?”). Conventional typing may exclude a person from a crime or a paternity suit, but it cannot, with any degree of certainty, prove that a particular person is the person whose bodily fluid was analyzed. And it couldn’t prove, to the satisfaction of the British Home Office, the mother and son relationship between Mrs. Sarbah and Andrew, particularly since the father was not available for testing. She could equally well have been Andrew’s aunt, rather than his mother (she did in fact have several sisters in Ghana).
You can imagine the distress in the Sarbah family, a feeling shared by members of The Hammersmith Law Centre. At that point, one of the Centre’s lawyers read a newspaper article about some scientific research that sounded potentially helpful. The article featured the work of Dr. Alec Jeffreys of Leicester University. It described how his research group had recently discovered that human DNA carried certain patterns that were highly variable in the population (3, 4). In fact, Dr. Jeffreys was of the opinion that they might be so variable that they could provide individual-specific DNA ‘fingerprints’. Although there were still only limited statistical data to support his idea, he was getting more evidence in support of it every day. The members of the Law Centre wondered whether the familial relationship of Mrs. Sarbah and her son could be established using Dr. Jeffreys’ approach, and so they contacted him. He was a fairly recent faculty appointment to the university, but had a solid pedigree in research. If he thought it merited a chance, why not try it?
Dr. Jeffreys had not done anything remotely like what he was now being asked for. Nevertheless, he immediately agreed to carry out his DNA analysis on blood samples from Mrs. Sarbah, her three undisputed children, and Andrew. The results made history (2). Indeed, all of the family members could be linked to each other through their DNA patterns, even without a sample from the father. The enlightened Home Office decided that Dr. Jeffreys’ test provided sufficient proof of the claimed relationship between Mrs. Sarbah and Andrew. It was the first time that such a test had been used in a legal case, and it was so interesting scientifically that it was reported in the journal ‘Nature’, one of the most prestigious research journals in the world (5). What Dr. Jeffreys’ results showed was that it is possible to prove a parent – child relationship beyond a reasonable doubt, even when the second parent is not available. The field of DNA profiling, the name that was adopted to avoid confusion between DNA fingerprinting and the more conventional kind, was born.
In the words of Dr. (now Sir Alec) Jeffreys, the floodgates opened, and he and his part-time technician were swamped by requests for DNA testing, since initially they were the only group able to do it (1). In addition to scrambling to fill the need, they began training others to use it, and it was soon introduced into a number of laboratories in England and around the world to accommodate the demand. And law enforcement agencies pounced on the new technology.
From Blood Relatives to Bloodstains
Dr. Jeffreys then tested whether old biological samples, such as dried bloodstains, would provide useful material for DNA profiling (6). They did. The technique would work on such samples, although in its original form it required a fairly large quantity of material (in time, the sensitivity was increased dramatically, as described in the next post). The Jeffreys group soon had an opportunity to test this kind of application in a real crime case. There had been two rape-murders in a small nearby village. The police had a suspect who had confessed to one of the crimes, but not the other. They thought that he had probably done both, based on their similarities. To try to link the other crime to their suspect, they asked Dr. Jeffreys to apply his technique to the evidence. He agreed, even though it was the first time for such an application, and the case had a high profile; failure might have been devastating. This led to a surprising, if convincing, outcome, as described in an interview with Dr. Jeffreys in 2009 (1):
The forensic samples arrived, and I have to say that was a chilling moment. [You’re] An ordinary academic and suddenly you’ve got murder samples in front of you. I remember my blood literally running cold at that point. We put the first probe on, and the prime suspect wasn’t a match (with the semen sample from the second murder)! Suddenly we were into the world of exclusion, and how many probes do you need for that? One. The result was so wacky, so totally out of keeping from what the police were expecting to see. We thought better do another one (probe). The results were totally astonishing, totally overturned what the police had got fixed in their minds about the guilt of this prime suspect. He was released.
The police said, OK we now believe all this DNA testing, let’s go and pan the entire local community and see if we can flush out the true murderer. . . The upshot of that was that the true perpetrator was flushed out, and the rest is history.
Incredibly, the first suspect had confessed in custody to one of the murders, but the DNA evidence showed that the same person had done both, and it wasn’t him. Indeed, subsequent experience has shown that, strange to say, confessions, like other evidence, are often not trustworthy.
A New Tool For Genetic Identification
The structural feature of DNA that Jeffreys and others had identified can be illustrated with a simple example. In the genomic DNA sequence of the first person in the figure below, there are 5 repeats of the sequence motif GCTTA (and its complement) between the positions N1 and N2 at a particular point in the genomic DNA. (The genomic sequences complementary to N1 and N2 are denoted N1′ and N2′).
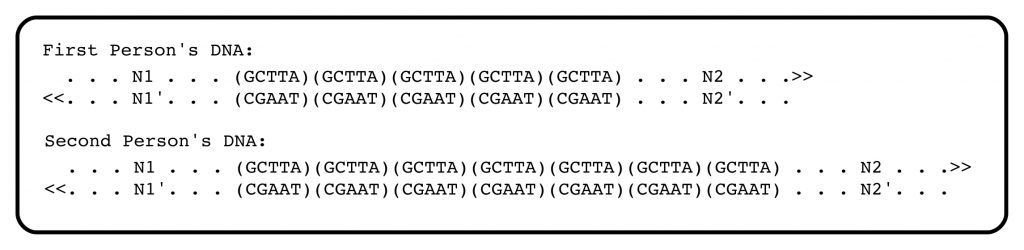
The second person has seven of the tandemly arranged, short sequences. If the distance between flanking sequences N1 and N2 can be measured accurately, the DNAs from these two people will be distinguishable. The flanking sequences N1 and N2 are unique in the genome, and measuring the distance between them, and thus the length of the tandemly-repeated sequence motif, is at the heart of DNA profiling. Within human (and other) populations, the numbers of such repeated short sequences do, in fact, often differ.
The Unexpected Discovery of ‘DNA Scissors’
In the original method, N1 and N2 were sites in the DNA that were cleaved by a special kind of enzyme, a “restriction endonuclease”. Their discovery had preceded the invention of DNA cloning, indeed, was critical for that invention. It all began with an unexpected result, published in the “Journal of Molecular Biology” in May 1970. I can still remember the frisson when I opened our copy of the journal and saw those papers. I was standing next to the sink closest to the door. The restriction endonucleases cleave DNA at very specific sites, when they recognize and attack a specific, short sequence motif (no relation to the short sequence in the VNTR region). So genomes (both human and nonhuman) could now be reproducibly cleaved into precise fragments of various lengths. It was instantly obvious that such a discovery changed everything.
Human DNA contains about 3.2 billion base pairs per haploid genome. It can be randomly cleaved by physical abuse (actually, no matter how gently a sample of human DNA is handled, it’s impossible not to cleave it), or by enzymes that randomly attack the DNA backbone. Such fragments are random, and not useful for any serious analysis. But with the discovery of restriction enzymes, DNA could be cleaved into reproducible, bite-sized pieces.
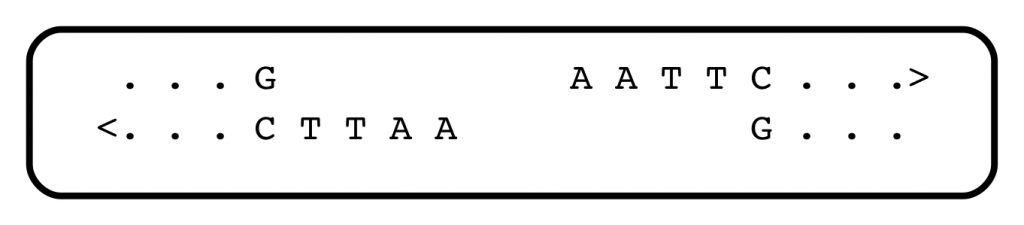
There soon followed a deluge of discoveries of restriction endonucleases — there are hundreds of them, each recognizing different short sequence motifs, in the world of bacteria (some of them, from different sources, have the same specificities). Almost all recognize a short sequence with internal symmetry, and cleave the DNA within that sequence. They serve as a primitive kind of immune system against infection with foreign DNA. One heavily-used restriction endonuclease, EcoRI (the names are bizarre, and reflect their cells of origin), recognizes and cleaves the string GAATTC:
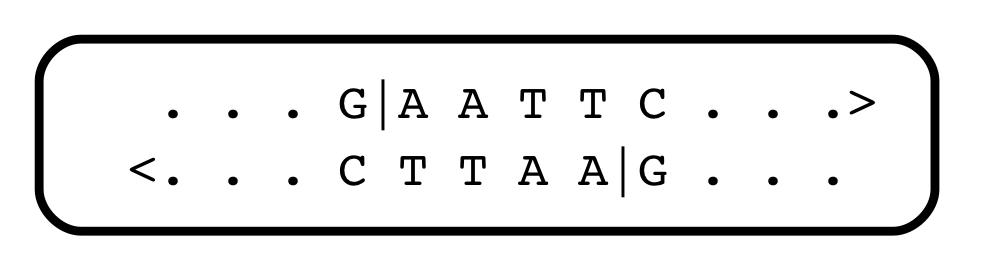
Jeffreys could analyze VNTRs using one or more restriction enzymes. The distances between N1 and N2, which were sites of cleavage by restriction enzyme(s), could be determined by electrophoresis, which separates DNA fragments on the basis of length. It soon became apparent that the family of DNA fragments produced by cleavage with restriction enzymes, and containing one or another VNTR, was an extremely personal “fingerprint”, and the science of DNA profiling entered the courtroom, where it has flourished.
As for the restriction enzymes themselves, soon after they were discovered, they opened Pandora’s box: Molecular cloning. That’s another fascinating story, but suffice to say that most restriction enzymes generate physical ends of fragments that associate with each other, randomly. Look at the Eco RI cleavage site above: after the enzyme does its work, every fragment of DNA ends with the same sequence:
The ends of all fragments have a complementarity, and can find each other and stick together, albeit transiently, because of that. There exist enzymes that can stitch them back together during their brief interaction. Because every EcoRI fragment ends with the same, complementary, sequence, the rejoining of fragments is random. Human DNA cleaved with EcoRI can be stitched into bacterial DNA also cleaved with EcoRI. Together with the ability to re-introduce DNA into living cells, this meant that recombinant DNA could be generated and cloned. Which allowed for the isolation of globin DNA, and the discovery of VNTRs, a crowd favourite for CSI.
Sharpening the Tools
DNA profiling was shown to be a reliable technique almost as soon as it was invented, with greater discriminatory power than existing approaches. But at first, it was not very sensitive. As described in the following post, DNA profiling was about to become massively more sensitive, a development that made it more powerful, but which also opened the door to some concerns.
Go to Latest Posts
Sources cited
- Gitschier, J. 2009. The Eureka moment: An interview with Sir Alec Jeffreys. PLoS Genetics 5:1-4.
- Millard, J. 1985. That’s my boy. Mother turns to science in fight for son. In Shepherd’s Bush Hammersmith Gazette, Friday, May 17, 1985 (available at http://www.dnai.org/d/index.html).
- Jeffreys, A. J., V. Wilson, and S. L. Thein. 1985. Hypervariable ‘minisatellite’ regions in human DNA. Nature 314:67-73.
- Jeffreys, A. J., V. Wilson, and S. L. Thein. 1985. Individual-specific ‘fingerprints’ of human DNA. Nature 316:76-79.
- Jeffreys, A. J., J. F. Y. Brookfield, and R. Semeonoff. 1985. Positive identification of an immigration test-case using human DNA fingerprints. Nature 317:818-819.
- Gill, P., A. J. Jeffreys, and D. J. Werrett. 1985. Forensic application of DNA ‘fingerprints’. Nature 318:577-579.